Jim LeNomenclatoriste
Je suis un mignon petit Traquet rubicole

Maybe it's just a typo

Maybe it's just a typo

This may not fit in perfectly here, but I don't know where else to put it, and given the fact that the contributors in this thread are highly interested in ducks and geese I might have a chance to get a qualified input to the issue here.
I shot the male wigeon in Denmark in December 2017 with the extra black bar on the wing (the right hand wing).
Wonder if anybody has seen this before? I never have, but got the idea that it could be a special feature in a part population somewhere. Any comment will be welcome.
By the way, I can't open the link that opens the thread here as it needs a pass word !


I am not looking for a "hunting forum" - I am just trying to find out, whether this unusual wing is a random mutation or is typical of some part population somewhere. Please tell me if there should be a better forum in here to get an answer. I just thought taxonomy would be the closest I could get to get an answer as it has a lot to do with plumage.





Proposal (825) to SACC
Treat Sarkidiornis sylvicola as a separate species from Sarkidiornis melanotos

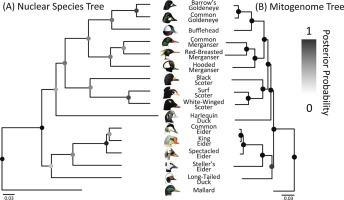
So am I...Lavretsky P., Wilson R.E., Talbot S.L. & Sonsthagen S.A. (2021). Phylogenomics reveals ancient and contemporary gene flow contributing to the evolutionary history of sea ducks (Tribe Mergini)
I'm also interested by this paper

You'll be. With your mail in pmSo am I...
MJB
and me tooSo am I...
MJB

Give me your mail by PM 😊Could someone send this over to me too, please. Cheers,
