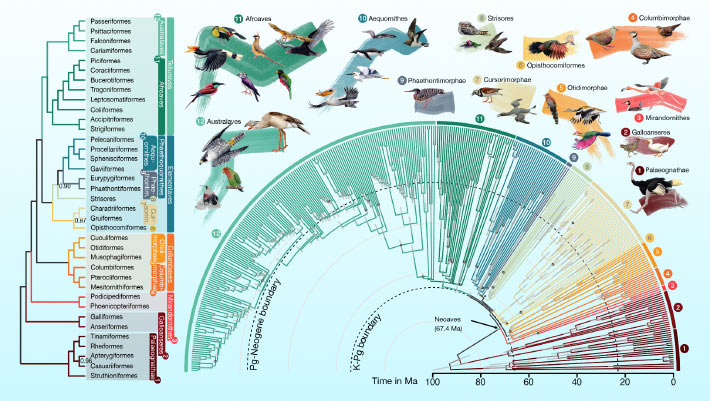
Massive family-level global bird tree based on whole-genome analyses
BOOM! Today, our new bird phylogeny was published in
Nature (soon also at
https://doi.org/10.1038/s41586-024-07323-1). Or, well, an ugly preview version that should soon be prettified was made available. This is the main result of the family phase of the
Bird 10,000 Genomes Project (B10K), and thus is “the new Jarvis et al. (2014) tree”. The work has been incredibly intensive and demanding, and the 52-headed crew of co-authors has been led by Josefin Stiller (University of Copenhagen), Siavash Mirarab (UCSD) and Guojie Zhang (Zhejiang University). I should stress already here that my contributions have been rather minor, but I’m probably the only one on BirdForum, so I figured it falls on me to report our study here.
This is the most detailed, highly resolved, and precise phylogeny of birds to date. It is based on 363 species, representing 92% of all bird families (as per Howard & Moore 4; the last 8% representing taxa we have so far been unable to get usable samples from), whose whole genomes have been sequenced, assembled, and annotated. They have then put into a whole-genome alignment and, in total, these analyses cover nearly 100 BILLION basepairs. With quite some margin, this makes up the most extensive phylogenetic analyses of birds to date. There’s long been a debate about whether increased taxon sampling is better to increase precision and reliability or increased genetic sequencing (more loci; more base pairs). Here, we’ve been able to test this. More about that below, but first some of the taxonomic news. Let’s start with something that makes some intuitive sense.
Strigformes and Accipitriformes are sister orders (but it’s complicated…)
In the main two previous genome-scale phylogenies by Jarvis et al. 2014 (based on 48 taxa and 48,000,000 bp of genes and ultra-conserved elements (UCEs)) and Prum et al. 2015 (based on 198 species and 395,000 bp from protein-coding genes), Strigiformes (owls) have been recovered as a sister lineage to a clade within Afroaves comprising Coliiformes, Leptosomatiformes, Trogoniformes, Bucerotiformes, Coraciiformes, and Piciformes, i.e. the bulk of clade Afroaves. Accipitriformes, however, was recovered as having branched off earlier from that clade (Jarvis et al. 2014) or even earlier, from the branch leading to the ancestor of Afroaves and the Australaves orders Cariamiformes, Falconiformes, Psittaciformes, and Passeriformes (Prum et al. 2015).
In our main tree, which was produced with coalescent-based analyses of a 63,430,000-bp matrix of spaced intergenic sequence (63,430 loci of 1,000 bp each), diurnal birds of prey and their nocturnal counterparts come out as sister lineages, placed basally within Afroaves. Having said that, these orders are particularly problematic to place and ancient hybridization may have played a role. More details in the paper. Now for a newly recovered grouping that none of you would have seen coming!
 Elementaves is an exceedingly diverse neoavian clade of surprising relatives
Elementaves is an exceedingly diverse neoavian clade of surprising relatives
As expected, at the base of our tree sits infraclass Palaeognathae (ratites), sister to all other extant birds, which sort under infraclass Neognathae. Neognathae diverges into parvclasses Galloanseres (fowl and ducks) and Neoaves, the latter comprising all remaining birds. The arrangement within parvclass Neoaves has been subject to many changes, since the very rapid diversification at its base has been near-impossible to sort out with any confidence. Our analyses recover four basal clades within Neoaves. The first one to branch out is Mirandornithes (Phoenicopteriformes, Podicipediformes); next comes Columbaves (Mesitornithiformes, Pterocliformes, Columbiformes, Musophagiformes, Otidiformes, Cuculiformes)—although there is a whole interesting story about Columbaves, that shows that Columbimorphae may in fact be sister to Mirandornthes instead, forming Columbea (see separate paper by Mirarab et al. released today in PNAS). Left is a two-way split between Telluraves, which comprises Afroaves and Australaves as above, and our novel cool gang/unruly bunch.
Included here is a clade with Opisthocomiformes as probable sister to the sister pair Gruiformes and Charadriiformes; another clade where Caprimulgiformes (sensu lato, i.e. = Strisores) likely sits at the base as sister to a clade which bifurcates into Phaetontimorphae (Eurypygiformes, Phaetontiformes) and Aequornithes (Gaviformes, Pelecaniformes, Procellariformes, Sphenisciformes). This means that our very smallest (hummers) and largest (one way to measure at least; albatrosses) are part of the same clade. I see lots of shaking heads and furrowed brows right now, and expect several of you to think that “this just cannot be right”. While no phylogeny can claim to be the truth, I will say, however, that this is not a random grouping that has just been accepted without challenge (and loads of additional analyses). It does nicely illustrate, however, how the amount of data affects both support and topology – and that different methods of analyses recover different topologies. There’s lots more to read about this in the paper.
As for the name, we name it Elementaves because the clade’s lineages have diversified into terrestrial, aquatic, and aerial niches (holding several of our most impressive specialists like penguins, albatrosses, and swifts), corresponding to the classical elements of earth, water, and air. So, what about fire? Well, several species within Phaethontimorphae have names derived from the sun, a big ball of fire.
What are the differences in topology and which tree should you believe in?
We argue overall that our extensive taxon sampling combined with extensive genomic sampling (i.e. not taxa vs loci, but preferably both) of regions that are not under direct or indirect selection (unlike previous trees), analysed in a coalescence-based framework, produces a stronger and more reliable bird phylogeny. There’s lots more to read about this in the paper. But what are the differences from the two previous main panavian phylogenies, Jarvis et al. (2014) and Prum et al. (2015)? This is made easy to assess as it is summarised in this figure:
View attachment EDF5annot_high.jpg
Should you trust this new tree over Jarvis or Prum? We argue yes, but we also offer a fun analyses that lends some support to this claim. If one explores the phylogenetic signal (based on the tree) in how morphological trait (co-)vary, it turns out that in morphometric variables (bill measurements, body mass, tail length, wing length and shape, and tarsus length), the distribution of measurements across species “make more sense” with the new tree. That is, there is less sudden variation (evolutionary shifts) along the tree branching toward the present with our new topology, than compared with the tree of Prum et al. (2015) [too few species in Jarvis et al. 2014].
Precision of age estimation and timing relative the
The final cool thing I’d like to bring up about this new phylogeny is that the precision is vastly higher for node age estimation than has ever been achieved before (including Jarvis et al. 2014 and Prum et al. 2015). This has much to do with the extensive work to classify avian fossils and empirically generate ‘calibration densities’ for no less than 34 nodes. Many node age estimates differ from previous studies, but most fall within the latter’s large 95% confidence intervals. Our new dating offers much narrower confidence intervals (and some surprises in terms of actual age, like very much older Tinamidae, older Apodidae, younger Pipridae and Accipitridae). This offers more insight to the timing around the Cretaceous–Paleogene boundary, which coincides with the rapid and extensive diversification of Neoaves. Some folks have proposed a scenario in which the main lineages of Neoaves would have evolved prior to the asteroid impact and survived through it. More favoured and more well-supported by fossils, though so far not supported with any precision by molecular data, is the ‘big bang’ scenario. This means that the main diversification of Neoaves would have happened after the asteroid impact, as a response to the vacation of whole ecological niches previously occupied by non-avian dinosaurs and other animals. Our tree lends support to this scenario.
That’s over and out from me at this point, but I promise you that there is much more to be gleaned from Stiller et al. (2024)! Not least, a lot of additional goodies are hiding in the supplemental extended data figures, so don’t miss out on them.